Preliminary assessment of the genetic diversity of Ecuadorian Ilex guayusa using inter simple sequence repeat (ISSR) markers: Genetic diversity of Ecuadorian Ilex guayusa
Estudio preliminar de la diversidad genética de Ilex guayusa ecuatoriana mediante marcadores inter simple sequence repeat (ISSR)
ACI Avances en Ciencias e Ingenierías
Universidad San Francisco de Quito, Ecuador
Received: 09 June 2016
Accepted: 30 December 2017
Resumen: Evaluamos el grado de diversidad genética de Ilex guayusa, una especie de árbol de importancia etnobotánica y comercial para las comunidades indígenas de la Amazonía ecuatoriana. Caracterizamos genéticamente a 157 individuos, provenientes de chacras ubicadas a lo largo de seis provincias, utilizando nueve marcadores ISSR (Inter Simple Sequence Repeats). Aunque se detectó un total de 91 bandas polimórficas a lo largo del set completo de muestras, el índice de heterocigosidad estimada (He = 0.19) reveló un nivel reducido de variabilidad genética para la especie. El análisis de varianza molecular (AMOVA) demostró que el 82% de la variación observada para I. guayusa ocurrió dentro de poblaciones, y solo el 18% entre poblaciones. Los índices de distancias genéticas de Nei (0.013 < Ds < 0.086) revelaron un nivel reducido de divergencia genética entre individuos provenientes de provincias diferentes. El bajo grado de diversidad genética encontrado para I. guayusa puede ser atribuido al hecho de que esta especie es cultivada exclusivamente mediante propagación clonal; una actividad cultural que probablemente ha homogenizado el acervo genético de la especie a lo largo de su rango geográfico de cultivo. El análisis de coordenadas principales (PCoA) demostró que el germoplasma colectado puede estructurarse en tres grupos distintos caracterizados por un leve contraste genético en gradiente direccional, de norte a sur. La inclusión de un mayor número de muestras provenientes de provincias sub-representadas (ej. Sucumbíos) y poblaciones silvestres, si existen, ayudaría a resolver los vacíos en el conocimiento respecto a la diversidad genética y estructura poblacional de la especie y la historia de su cultivo en la región.
Palabras clave: Aquifoliaceae, Guayusa, propagación clonal, baja diversidad genética.
Abstract: We assessed the degree of genetic diversity of llex guayusa, a tree species of ethnobotanic and commercial relevance for indigenous communities of the Ecuadorian Amazon. We characterized 157 individuals, from small cultivation sites across six provinces, using nine Inter Simple Sequence Repeat (ISSR) markers. A total of 91 polymorphic bands were detected across the complete sample-set, but estimated heterozygosity (He = 0.19) revealed a reduced level of genetic variability. Partitioning of genetic diversity (AMOVA) indicated that 82% of the variation observed for I. guayusa occurred within populations, and only 18% between populations. Pairwise- Nei genetic distance indices (0.013 < Ds < 0.086) implied a reduced level of genetic divergence between individuals from different provinces. The low degree of genetic diversity found for I. guayusa could be ascribed to the fact that the species is exclusively cultivated via clonal propagation; a cultural activity which has probably homogenized the species’ genetic pool across its geographic range of cultivation. However, PCoA analysis resolved collected germplasm into three distinct groups displaying a subtle genetic contrast in a directional gradient, from north to south. The inclusion of a higher number of samples from underrepresented provinces (i.e. Sucumbíos), and wild populations, if they exist, would help resolve knowledge gaps regarding the genetic diversity and population structure of the species and its cultivation history in the region.
Keywords: Aquifoliaceae, Guayusa, clonal propagation, low genetic diversity.
INTRODUCTION
Ilex guayusa Loes. (Aquifoliaceae) is a tree species native to the Amazonian rainforests of northern South America [1]. In Ecuador, it is almost exclusively found in small cultivation sites (commonly known as chacras) along the western basin of the Amazon. It currently represents the most harvested and widely utilized non-food plant species by indigenous groups, like the Kichwa, Achuar and Shuar [2,3]. These ethnic groups typically use the leaves of I. guayusa to prepare ceremonial beverages conferring good luck for hunting and fishing and for protection against snake bites [2-5]. Ilex guayusa leaves are also employed for ritualistic purging, and to treat ailments like gastritis, stress and infertility [5,6]. The leaves of I. guayusa are rich in caffeine, terpenes, phenols and other methylxanthines, and exhibit putative antiinflammatory, microbicidal, anti-oxidant and anti-obesity properties [4,5,7].
Historically, dried I. guayusa leaves were only sold in low volumes as a specialty product in local markets throughout the Ecuadorian Amazon. However, since 2009 and principally owing to the efforts of RUNARM, a social enterprise based in the USA and Ecuador, I. guayusa has gained commercial notoriety in international markets. It is commercialized as soluble teas and canned drinks with energizing and stimulant properties, mainly in the United States and Canada. New export markets have thus generated an increase in the number of I. guayusa producers in the Ecuadorian Amazon, from approximately 300 in 2011, to over 1,600 in 2013 [8]. Certainly, the growth in popularity of I. guayusa can bring socio-economic change to the region, but further expansion in its commercial production must be carefully evaluated as the species is native to a fragile and highly biodiverse ecosystem, vulnerable to the detrimental effects of intensive monocropping [8].
Despite its rich ethnobotanic relevance and rising commercial prospects, the biology, cultivation history and genetics of Ilex guayusa are poorly understood. This research presents a first assessment of the genetic diversity of I. guayusa in the Ecuadorian Amazon using Inter-Simple Sequence Repeat (ISSR) markers. This preliminary survey of the degree and geographic distribution of the genetic diversity of the species is necessary for establishment of diversity conservation strategies, design of plant improvement programs and generation of sustainable- agroforestry systems that best utilize plant genetic resources in vulnerable ecosystems.
MATERIALS AND METHODS
Plant material and DNA isolation
RUNA’s technicians, in collaboration with students from USFQ, collected leaf samples from 157 Ilex guayusa individuals, from small cultivation sites listed in their database of I. guayusa leaf providers. The sites were distributed across the provinces of Sucumbios (1), Napo (91), Orellana (7), Pastaza (35), Morona Santiago (11) and Zamora Chinchipe (12); all located in the Ecuadorian Amazon (Fig 1). Collected samples were georeferenced using a Garmin E-Trex Legend HCx GPS system (Garmin International Inc., USA). For every individual, young and healthy leaves were collected from the lower branches of trees and preserved at -20°C. Genomic DNA was isolated from leaf samples using the CTAB procedure described by Kieleczawa [9]. Following extraction, DNA quality and concentration were assessed using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, USA). DNA samples were diluted to a final concentration of 20 ng/gl.
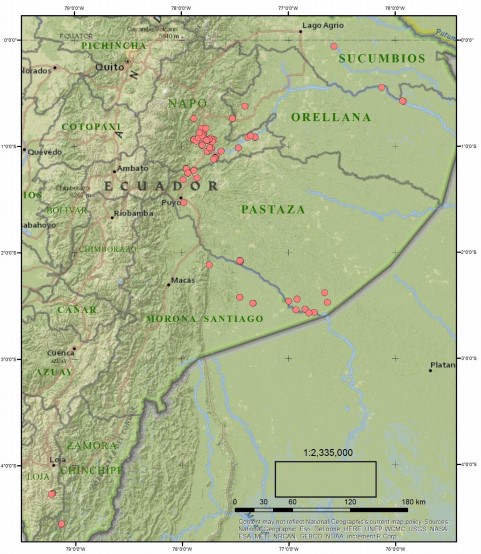
Figure 1
Location of the 157 I. guayusa individuals collected marked as pink circles.
PCR amplification
Nine anchored ISSR primers were used for the molecular characterization of Ilex guayusa (Table 1). These primers belong to the universal UBC ISSR primer set (University of British Columbia, Canada) and had been successfully employed by Zhou [10] for the genetic characterization of diverse Ilex species. Genomic DNA (40 ng) was PCR amplified in a 20 gl reaction containing 50 mM KCl, 20 mM TrisHCl (pH 8.4), 1 gg/gl BSA, 2.0 mM of MgCh, 0.16 mM Primer Mix, 0.5 mM dNTPs, and 2 units Taq Polymerase (Life Technologies, Carlsbad, California). PCR amplification conditions consisted of an initial denaturation at 94°C for 4 min; followed by 40 cycles of a 1 min denaturation at 94°C, annealing of 50 sec at 52°C, an extension at 72°C for 1 min; and a final extension period of 7 min at 72°C. All reactions were performed in a T-Personal Series Thermocycler (Biometra, Goettingen, Germany). Amplified products were separated and visualized via electrophoresis in 1.75% w/v agarose gels, run at 90 V for 2 hours, and stained with SYBR Safe DNA gel stain (Life Technologies, Carlsbad, California). Gel band sizes were estimated using a 1 Kb DNA ladder (Life Technologies, Carlsbad, California) and band scoring was performed visually based on band-size differentiation. All bands were scored as presence/absence markers, and a binary raw-data matrix, where the presence of a band was encoded as one (1) and the absence of a band as cero (0), was generated for all gel banding patterns.
| Primer code | Primer sequence | Number of amplified bands | Number of polymorphic bands | Polymorphism percentage |
| UBC-815 | (CT)8G | 11 | 10 | 90.91 |
| UBC-824 | (TC)8G | 7 | 6 | 85.71 |
| UBC-825 | (AC)8T | 15 | 13 | 86.67 |
| UBC-826 | (AC)8C | 12 | 12 | 100.00 |
| UBC-827 | (AC)8G | 15 | 14 | 93.33 |
| UBC-840 | (GA)8YT | 11 | 10 | 90.91 |
| UBC-846 | (CA)8RT | 11 | 7 | 63.64 |
| UBC-857 | (AC)8YG | 18 | 16 | 88.89 |
| UBC-860 | (TG)8RA | 6 | 3 | 50.00 |
| Total | 106 | 91 | 85.85 |
Information of nine ISSR anchored primers used for the molecular characterization of 157 Ilex guayusa individuals from the Ecuadorian Amazon.
Data analysis
For all identified loci expected heterozygosity (He) was determined using the GenAlEx 6.501 software package [11]. Accordingly, within- and between-population estimates of molecular variance (AMOVA) and Nei genetic diversity indices were obtained using GenAlEx 6.501; assuming individual provinces as independent genetic groups, and excluding Sucumbios because only one sample was available for this province. Principal coordinates analysis (PCoA) was performed with DARwin 5.0.15 using the DICE genetic dissimilarity coefficient [12].
RESULTS
Collectively, all evaluated ISSR markers yielded a total 91 polymorphic bands across the complete sample set (157 individuals). The total number of polymorphic loci scored per primer ranged from 3 (UBC-860) to 16 (UBC-857), with an average of 10.1 loci per primer. Of the 91 bands analyzed, 73 polymorphic loci occurred in low frequencies (<0.5%) across the sample-set, but none were exclusive to any given province or genotype group. The expected heterozygosity estimate for Ilex guayusa (He = 0.19) revealed a low degree of genetic diversity for the collected germplasm. Accordingly, pairwise-Nei genetic distance indices implied a reduced level of genetic variability for this species (Table 2). The greatest degree of genetic differentiation was detected between individuals from Napo and Zamora Chinchipe (Ds = 0.086); the two most geographically distant provinces. The lowest genetic distance was detected between individuals from the neighboring provinces Napo and Orellana (Ds = 0.013). While a Mantel test did not reveal a significant association between genetic and geographic distances (r=0.060; p=0.037), geographically distant provinces generally displayed a higher degree of genetic divergence.
| Napo | Orellana | Pastaza | Morona Santiago | Zamora Chinchipe | |
| o.ooo | Napo | ||||
| 0.013 | 0.000 | Orellana | |||
| 0.040 | 0.032 | 0.000 | Pastaza | ||
| 0.040 | 0.023 | 0.025 | 0.000 | Morona Santiago | |
| O.O86 | O.O75 | 0.044 | 0.027 | o.ooo | Zamora Chinchipe |
Nei genetic distance matrix comparing Ilex guayusa individuals from different provinces in the Ecuadorian Amazon. Sucumbios was excluded from the analysis as only one sample was available for this province.
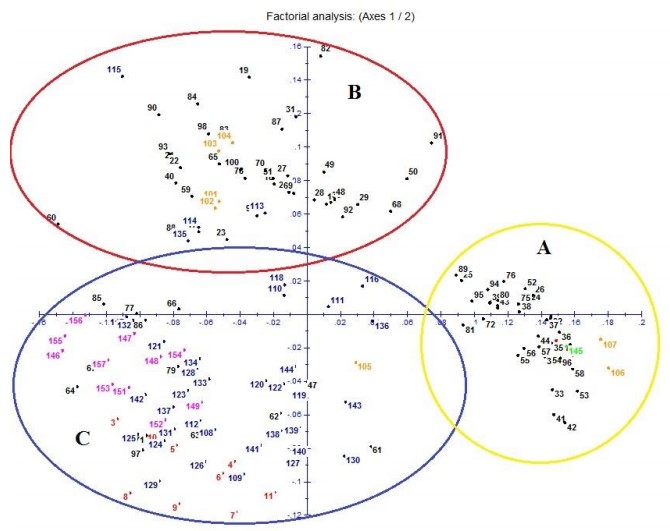
Analysis of molecular variance (AMOVA) demonstrated that the highest percentage (82%) of genetic variation for Ilex guayusa was found within groups (individuals belonging to the same province), relative to among groups (18%). This factor indicates a low degree of population differentiation. Nevertheless, Principle Coordinates Analysis (PCoA) (Fig 2) could resolve collected germplasm into three robust genetic clusters as determined by its first two coordinates, each respectively accounting for 23% and 11% of observed variation. Cluster A included individuals from the most-northern areas of the provinces of Napo and Orellana, as well as the only individual from the province of Sucumbíos. Cluster B included individuals from central and northern quadrants of Napo, and individuals from the province of Orellana that were geographically proximal to Napo’s eastern provincial border. Finally, Cluster C comprised all individuals from the southern quadrants of Napo, and individuals from the southern Amazonian provinces of Pastaza, Morona-Santiago and Zamora-Chinchipe.
DISCUSSION
The main objective of this study was to assess the level of genetic diversity of Ilex guayusa, a species of great ethnobotanic and commercial importance for indigenous communities of the Ecuadorian Amazon [2]. Overall, expected heterozygosity (He = 0.19) revealed a reduced level of genetic variability for the species. For comparison, using five isoenzyme markers, Winge et al. [13] detected a comparatively higher degree of heterozygosity (He = 0.50) for Ilex paraguariensis, a close relative of I. guayusa. The RAPD-based characterization of four natural populations of I. paraguariensis from southern Brazil revealed that the average (Nei) genetic distance between individuals within a population was 0.392 [14], whereas the maximum degree of genetic differentiation between I. guayusa individuals analyzed in this study was 0.086 (Table 2). At first hand, the low level of genetic variability observed for Ecuadorian I. guayusa appears contradictory, especially when considering the extensive number of polymorphic loci (91) detected across the evaluated sampleset. However, it is important to highlight that approximately 80% of all detected polymorphisms occurred in low frequencies (<5%), and the vast majority of these did not exhibit correlative patterns of allelic fixation for specific genotypes or genotype groups (i.e. individuals from specific provinces).
The low degree of genetic diversity found for Ilex guayusa could most likely be ascribed to the fact that all collected germplasm originates from small cultivated plots. In the Ecuadorian Amazon, indigenous groups cultivate I. guayusa exclusively via clonal propagation through stem cuttings, a cultural activity that has probably homogenized the species’ genetic pool across its geographic range of cultivation [16-18]. Historically, I. guayusa has existed in close relationship with Amazonian ethnic and cultural processes, and the species’ sampled geographic distribution likely reflects the migration patterns of indigenous groups [15]. Moreover, no reports exist of wild I. guayusa populations, and it is therefore impractical to extrapolate on whether the species displays a generalized low degree of genetic diversity. The results of this study also imply that in the Ecuadorian Amazon, cultivated I. guayusa originates from a specific and reduced gene pool. Partitioning of genetic diversity (AMOVA) indicated that 82% of the variation observed for this species occurred within populations (provinces), and only 18% between populations. Natural populations of I. paraguariensis have also shown a reduced degree of genetic differentiation, principally accounted to the species’ outcrossing nature [14]. In outcrossing perennial tree species, natural populations generally exhibit a high degree of allelic exchange via gene flow; a factor which limits genetic divergence between populations [19]. Yet, because samples analyzed in this study come from cultivated plots established through clonal propagation, it seems probable that the high degree of genetic homogeneity observed for I. guayusa results from the widespread propagation of a selection of homogenous genotypes with desirable characteristics and established for cultivation prior to the anthropogenic-driven dispersion of the species across the Amazon basin [5].
Remarkably, while genetic differentiation between established groups was limited, a subtle genetic structure could be established for Ecuadorian Ilex guayusa. Principle Coordinates Analysis (PCoA) could resolve collected germplasm into three distinct groups. All clusters included a high proportion of individuals from the province of Napo, which was over-represented in this study. However, each cluster was distinctly and respectively characterized by the presence of individuals from the remaining northern, central and southern provinces of the Ecuadorian Amazon (Fig 2). For decades, Napo has represented the center of economic activity in the Ecuadorian Amazon, and around 50% of its population is composed of migrants from neighboring provinces. It is probable that some of these migrants brought specific variants of I. guayusa for cultivation [20]. Incidentally, PCoA clustering tended to group individuals from Napo in a directional gradient, from north to south, possibly highlighting the geographically structured introduction of novel I. guayusa alleles from distinct, yet proximal geographic locations in neighboring provinces. Certainly, the inclusion of a higher proportion of individuals from underrepresented provinces (i.e. Sucumbíos), as well as wild populations (if these should exist), would help us better resolve the geographic distribution of I. guayusa genetic diversity along the Amazon basin of Ecuador, and formulate stronger conclusions regarding its cultivation history in the region. This information will be crucial for the establishment of adequate conservation strategies aimed at preserving and promoting the use of unique sources of genetic variation which can increase the productivity and environmental resilience of cultivated orchards under sustainable agro-forestry systems. Expectantly, the latter will enable for a controlled and sustainable expansion of I. guayusa cultivation in the Ecuadorian Amazon which can meet rapidly growing commercial demands, and reduce the need for intensive mono-cropping systems and their associated detrimental effects on vulnerable ecosystems.
Acknowledgments
This project was funded in part by the support of the Inter-American Institute for the Cooperation in Agriculture’s Sustainable Forest Management Program and the Finnish Foreign Ministry. This research adhered to the legal requirements of Ecuador (Contrato Marco de Acceso a los Recursos Genéticos MAE-DNB-CM- 2016-0046). We would like to thank Eliot Logan Hines for all his support during the development of this research. We would like to acknowledge the RUNARM Foundation for their financial support and their active participation in the collection of Ilex guayusa samples. We would also like to extend our gratitude to all researchers from Plant Biotechnology Laboratory at Universidad San Francisco de Quito who have directly or indirectly contributed or participated in the development of the project.
References
Tropicos. (2016, August 4). Ilex guayusa Loes. Missouri Botanical Garden. Retrieved from http://www.tropicos.org/Name/2000086
Innerhofer, S., & Bernhardt, K. (2011). Ethnobotanic garden design in the Ecuadorean Amazon. Biodiversity Conservation, 20, 429-439. doi: http://doi.org/10.1007/s10531-010-9984-9
Bennett, C. (1992). Hallucinogenic plants of the Shuar and related indigenous groups in Amazonian Ecuador and Peru. Brittonia, 4, 483-493. doi: http://doi.org/10.2307/2807199
Crown, P. (2012). Ritual Black Drink consumption at Cahokia. Proceedings of the National Academy of Science, 109(35), 13944-13949. doi: http://doi.org/10.1073/pnas.1208404109
Lewis, W., Kennelly, E., Bass, G., Wedner, H., & Fast, D. (1991). Ritualistic use of the holly Ilex guayusa by Amazonian Jivaro Indians. Journal of Ethnopharmacology, 33, 25-30. doi: http://doi.org/10.1016/0378-8741(91)90156-8
Tene, V., Malagon, O., Finzi, P., Vidari, G., Armijos, C., & Zaragoza, T. (2007). An ethnobotanical survey of medicinal plants used in Loja and Zamora- Chinchipe, Ecuador. Journal of Ethnopharmacology, 111, 63-81. doi: http://doi.org/10.1016/j.jep.2006.10.032
Hao, D., Gu, X., Xiao, P., Liang, Z., Xu, L., & Peng, Y. (2013). Research progress in the phytochemistry and biology of Ilex pharmaceutical resources. Acta Pharmaceutica Sinica, 3(1), 8-19. doi: http://doi.org/10.1016/j.apsb.2012.12.008
Collen, W., Piñeiro, A., Krause, T., Logan-Hines, E. (Noviembre de 2013). Ilex guayusa como motor para el desarrollo sostenible en los sistemas agroforestales en la Amazonía occidental. En: Primer encuentro de bosques, recursos genéticos forestales y agroforestería. Encuentro realizado en el Instituto Nacional de Investigaciones Agropecuarias en Quito, Ecuador.
Kieleczawa, J. (2006). DNA sequencing II: optimizing preparation and cleanup. Vol. 2. Ontario: Jones & Bartlett learning.
Zhou, X. (2009). Inter-simple Sequence Repeat Molecular Markers in Ilex and the Tissue Culture System of Ilex cornuta Lindl Ex Paxt (Dissertation Thesis). Henan Agricultural University, Henan.
Peakall, R., & Smouse, P. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics, 28, 2537 2539. doi: http://doi.org/10.1111Zj.1471-8286.2005.01155.x
Perrier, X., & Jacquemoud-Collet, J. (2006). DARwin software. Retrieved from http://darwin.cirad.fr/
Winge, H., Wollheim, C., Cavalli- Molina, S., Assmann, E. M., Bassani, K. L. L., Amaral, M. B., & Mariath, J. E. A. (1995). Variabilidade genética em populates nativas de erva-mate e a implantado de bancos de germoplasma. Erva-mate: Biologia e Cutura no Cone Sul, 323-345.
Gauer, L., & Cavalli-Molina, S. (2000) . Genetic variation in natural populations of maté (Ilex paraguariensis A. St.-Hil., Aquifoliaceae) using RAPD markers. Heredity, 84, 647-656. doi: http://doi.org/10.10467j.1365-2540.2000.00687.x
Schultes, R. (1979). Discovery of an Ancient Guayusa Plantation in Colombia. Botanical Museum Leaflets, 27(6), 143-153. doi: http://doi.org/10.2307/41762818
Dueñas, J., Jarrett, C., Cummins, I., & Logan-Hines, E. (2016). Amazonian Guayusa (Ilex guayusa Loes): A Historical and Ethnobotanical Overview. Economic Botany, 20(10), 1-7. doi: http://doi.org/10.1007/s12231-016-9334-2
Van Der Hulst, R., Mes, T., Falque, M., Stam, P., Den Nijs, J., & Bachmann, K. (2003). Genetic structure of a population sample of apomictic dandelions. Nature Heredity, 90, 326335. doi: http://doi.org/10.1038/sj.hdy.6800248
Pluess, A., & Stocklin, J. (2004). Population Genetic Diversity of the Clonal Plant Geum reptans (Rosaceae) in the Swiss Alps. American Journal of Botany, 91(12), 2013-2021. doi: http://doi.org/10.3732/ajb.91.12.2013
Gottlieb, L. (1973). Enzyme differentiation and phylogeny in Clarkia franciscana, C. rubicunda and C. amoena. Evolution, 27, 205-214. doi: http://doi.org/10.2307/2406961
Del Popolo, F., Oyarce, A., Ribotta, B., & Rodriguez, J. (2007). Indigenous peoples and urban settlements: spatial distribution, internal migration and living conditions. Vol. 78. Chile: United Nations Publications. Retrieved from http://repositorio.cepal.org/bitstream/handle/11362/7219/S0700810_en.pdf?sequence=1
Notes