Pseudomonas aeruginosa transition from environmental generalist to human pathogen
Pseudomonas aeruginosa transición de generalista ambiental a patógeno humano
ACI Avances en Ciencias e Ingenierías
Universidad San Francisco de Quito, Ecuador
Received: 07 March 2021
Accepted: 04 May 2021
Abstract: Opportunistic bacteria Pseudomonas aeruginosa is a major concern as an etiological agent of nosocomial infections in humans. Many virulence factors used to colonize the human body are the same as those used by P. aeruginosa to thrive in the environment, such as membrane transport, biofilm formation, oxidation/reduction reaction, and others. The origin of P. aeruginosa is mainly from the environment; the adaptation to mammalian tissues may follow a source-sink evolution model. The environment is the source of many lineages, some of them capable of adaptation to the human body. Some lineages may adapt to humans and go through reductive evolution in which some genes are lost. The understanding of this process may be critical in order to implement better methods of controlling outbreaks in hospitals.
Keywords: bacteria, host adaptation, evolution, opportunistic.
Resumen: La bacteria oportunista Pseudomonas aeruginosa es una de las principales preocupaciones por su rol como agente etiológico de infecciones nosocomiales humanas. En muchos casos, los mismos factores de virulencia que le permiten a P. aeruginosa causar infecciones en humanos pueden ser empleados para prosperar en el ambiente, como los sistemas de transporte de membranas, la formación de biopelículas, las reacciones de oxidación / reducción y otros. El origen de P. aeruginosa es principalmente el ambiente; mas su adaptación a los tejidos de los mamíferos puede seguir un modelo de evolución del tipo fuente-sumidero. El ambiente es la fuente de muchos linajes de esta bacteria, donde algunos de ellos son capaces de adaptarse al cuerpo humano. Algunos linajes pueden adaptarse a los humanos y pasar por una evolución reductiva en la que se pierden algunos genes. La comprensión de este proceso puede ser fundamental para implementar mejores métodos de control de brotes en los hospitales.
Palabras clave: bacteria, adaptación al hospedador, evolución, oportunista.
INTRODUCTION
Pseudomonas aeruginosa is a Gram-negative aerobic rod bacteria, ubiquitous in the environment, and an opportunistic human pathogen [1]. It accounts for 9-19% prevalence of bacterial nosocomial infections and 7% of community-acquired pneumonia cases [2, 3]. Pseudomonasa eruginosa infections occur in the respiratory tract [4,5], eyes [6], ears, skin wounds [7], bloodstream [1], or surgical site infections [8]. Pseudomonas aeruginosa can colonize human intestines and skin, and it can take advantage of any host’s immunodeficiency to produce acute or chronic systemic infections [4, 9]. Moreover, it is argued that P. aeruginosa infections are acquired from the bacterial population that colonizes the proximal environment of the host [10].
Pseudomonas aeruginosa prospers in soil, tap water, plants, intestinal contents, food, puddles, and swimming pools; it has some remarkable abilities to survive in the presence of heavy metals and chlorine [11]. Pseudomonas aeruginosa lives surrounded by predatory amoeba, which may have selected bacterial cells with the ability to survive phagocytosis [12]. Living outside the host, P. aeruginosa adapts to changes (physical and chemical) and competes with other environmental microorganisms. This bacterium is a generalist and heterotrophic, possessing an arsenal of enzymes to oxidize many organic carbon sources, mostly decaying organic matter (and even xenobiotic compounds), to obtain energy [13-19].
One of the main problems associated with P. aeruginosa is the occurrence of nosocomial outbreaks where the source of the bacteria is unknown; many times hospitals have resorted to extreme measures such as plumbing system replacements to stop P. aeruginosa dissemination [20, 21]. Also, multidrug resistance P. aeruginosa is one of the most critical concerns of hospital-acquired infections [8].
In this review, we describe the complexity of the evolutionary processes involved in the adaptation of environmental P. aeruginosa to human tissues. The sporadic infections by environmental strains and infections by strains adapted to humans are examined. We will not address antibiotic resistance which is comprehensively covered by other manuscripts [22-24].
SPORADIC INFECTIONS BY ENVIRONMENTAL STRAINS
Pseudomonas aeruginosa infections with environmental strains involve the migration of bacteria from a heterogeneous ecosystem (environment) to a more homogeneous and restrictive ecosystem (human tissues). This movement is also known as a sourcesink dynamic. The source is the environment to which the bacteria are adapted, and the sink is the human tissue that is often a harsher milieu. The bacterial growth rate in the sink may not compensate for the death rate; therefore, the bacterial population in the sink is maintained by a constant introduction of bacteria from the source (environment outside the host) [25].
The adaptation of P. aeruginosa to different environments implies multiple mechanisms of DNA modifications such as inheritance of mutations, homologous recombination, horizontal gene transfer (acquisition of accessory genome), and gene deletion [26, 27]. Environmental strains of Pseudomonas obtain nutrients from human tissues and neutralize immune responses using the same genes that are useful for dealing with environmental challenges [14, 28, 29]. For example, a type 3 secretion system (T3SS) is a translocation apparatus enabling the bacteria to export effector proteins from the bacterial cell to a eukaryotic cell without an extracellular step. Effector proteins can cause different consequences in the eukaryotic cell; exoenzyme (Exo) U is a phospholipase that induces cell death of predatory amoebas [28, 30] whereas ExoS, another T3SS-exported effector protein, is involved in anti-predatory responses against free-living amoeba [31]. The same T3SS and its ExoU phospholipase also kill macrophages [32]; the effector protein, ExoS, has two domains that act in ADP-ribosylation and GTPase activities in host cell proteins [32] and also activates Toll-like receptors in phagocytes [33].
In the environment outside the host, stress caused by toxic levels of metal ions like copper, iron, and zinc induces the synthesis of pyoverdine, superoxide dismutase, fumarate hydratase, metal cation efflux transporter CzrA, ATP-binding cassette transporters (ABC transporters), copper resistance oxidase proteins, and others [34]. These molecules protect P. aeruginosa from oxidative insults and increase the availability of iron. In the host, the same molecules protect bacteria against oxidative reactions (from neutrophils or macrophages) and help to capture iron [35], as iron is sequestered in the host’s proteins [36]. Iron associates and inactivates Fur-like transcriptional repressors, which downregulate many genes required to scavenge iron from animal tissues [37].
The range of niches in different environments outside of humans is reflected in vast P. aeruginosa diversity [38]; this diversity makes it challenging to identify the environmental source of clinical strains, and some researchers indicate that all environmental P. aeruginosa could cause human disease [39]. In the last years, whole-genome sequencing (WGS) provided irrefutable evidence of the environmental origin of clinical P. aeruginosa [26, 27, 40, 41]. Some environmental clones are genetically indistinguishable from clinical isolates [39]; recently, whole-genome sequence comparison of geographically related strains (belonging to ST-1146, Mallorca, Spain) obtained from the environment and a clinical case showed no genetic differences, although the clinical isolate had a mutation in the oprD gene (causing carbapenem resistance) [42].
Nevertheless, recent evidence suggests also that not all environmental P. aeruginosa strains may be able to cause human infections; apparently some phenotypes are required to become human pathogens [43, 44]. Some evolutionary steps enabling P. aeruginosa strains to colonize human tissues may have occurred in the environment outside the host [27]; selective forces in niches outside the host may simulate some conditions in host tissues. For example, the abundance of environmental amoeba may select bacterial lineages able to survive macrophage attack. Nucleotide polymorphisms along the genome of clinical isolates (including epidemic and non-epidemic CF strains) have signs of positive selection (high rates of non-synonymous mutations, dN/ dS > 1) [26, 45]; among the genes under positive selection are those involved in LPS biosynthesis, flagella, secretion systems, iron scavenging, and iron uptake [26]. Analysis of P. aeruginosa genomes also shows that strains causing clinical infections have suffered higher levels of homologous recombination in genes similar to those showing signs of positive selection: membrane transport structures, biofilm formation, oxide/reduction, and other cell wall functions [27]. The recombination in the genome regions where positive selection is observed and the coincidence of recombinations in the same genes in many clinical isolates suggest that natural selection is affecting these strains.
INFECTIONS BY STRAINS ADAPTED TO HUMANS
Recent evidence suggests that human-to-human transmission of P. aeruginosa may be associated with the ability of these strains to colonize intestines; patients suffering from P. aeruginosa infections were colonized by the same clones [46].
However, some P. aeruginosa may become adapted to humans and transmitted from person to person as happens among pulmonary strains of cystic fibrosis patients [47].
Human-adapted P. aeruginosa strains have suffered similar evolutionary pressures as strains causing sporadic infections; however, they have acquired additional adaptations during chronic infections, such as cystic fibrosis (CF) (Fig. 1). Pseudomonas strains, causing CF infections, tend to form a different group of P. aeruginosa populations from strains causing other types of infections in hospitals [27]. CF lung may select strains with additional aptitudes [26, 27] that may have been acquired in inhospitable niches outside the host before adaptation to human tissues [26, 27, 48, 49]. Some of these CF strains become epidemic strains (found in different continents and transferred from person to person) and show positive selection in core genome genes involved in oxidation/ reduction, membrane transport, and type III secretion systems, among others [26].
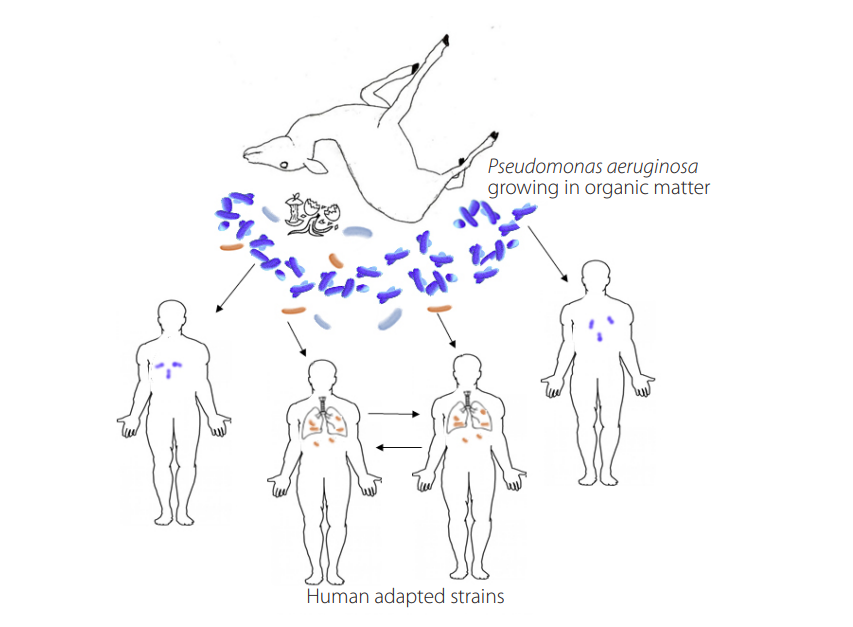
Figure 1.
Diverse Pseudomonas aeruginosa strains (blue bacteria) thrive in the environment degrading organic matter. Some strains have improved aptitude to grow in human tissue (brown bacteria); a subset of them have evolved additional adaptations and can be transmitted from human to human.
Going back to source-sink dynamics, if the sink conditions are not too harsh, the adaptation may occur especially if there is a constant migration rate [50] or in the presence of temporal less restrictive conditions [51]; chronic infections in CF patients may represent this. When an environmental generalist (such as P. aeruginosa) with a large genome (many genes required to thrive in an environment outside the host) [52-55] enters human tissue, a reductive evolution is prone to occur [18, 52-55]. In this scenario, evolution from environmental to pathogen involves gene loss due to small effective population size and lack of purifying selection; this gene loss is especially intense ifthese genes are unnecessary in the new niche (Fig. 1) [53, 56].
Longitudinal studies of cystic fibrosis P. aeruginosa strains have improved our understanding of evolutionary changes during chronic infections. Strains from these chronic infections show more profound evolutionary changes which probably occurred during infection of CF patients; paradoxically, some of these changes involve the loss of some apparent virulence factors. Evidence of reductive evolution may be apparent by the selection of amino acid auxotrophs in some strains [57, 58], loss of twitching motility [44], or reduction of type IV pili [59]; chronic infections of cystic fibrosis lungs may select patho-adaptive phenotypes [60]. Lungs from cystic fibrosis patients go through oxidative stress by the accumulation of hemoglobin, ferrous iron, and transferrin [61]. Pyoverdine (a siderophore involved in iron scavenging in human tissue) no longer seems a critical factor to obtain iron which may select pyoverdine mutants in genes such as pvdS (a sigma factor involved in pyoverdine transcription) and the pyoverdine gene and PrrF (an RNA that reduces pyoverdine expression and increases expression of HemO) [60]. An alternative iron-acquisition mechanism may take over in these mutants, allowing iron to be taken from the heme molecules; these strains may require heme oxygenase (HemO), which breaks the heme and releases biliverdin, carbon dioxide, and iron [62].
Also, the long-term colonization of the lungs selects variants of unusual biofilm formation. In P. aeruginosa, biofilm-mucoid phenotypes show increased expression of the exopolysaccharide PsI and alginate. PsI induction occurs by at least six different mutations in operons which occur when the infection lingers for a long time [63]. These strains also show mutations in the lasR gene, a transcriptional regulator of biofilm formation and other virulence genes [64].
Other P. aeruginosa adaptations to human hosts include higher mutation rates [65, 66], increased synthesis of multidrug-efflux pumps, higher antibiotic resistance, mucoid phenotypes, increased biofilm formation, higher tendency to form micro-colonies in tissue [67, 68], upregulation of metalloproteinases, lipid A modifications, reduced fucosyltransferase 2 expression, and better adhesion to tissues [27]. These host- adapted phenotypes have not been found outside the human host even in the same environment where infected patients reside [43, 44].
Some genotypes (LESA and ESB) could be found in CF patients on different continents [26, 27], suggesting human-to-human transmission. Evidence of human-to-human transmission has also been observed in some non-CF lineages (ST-111, ST-235, and ST-175) that caused infections in five hospitals in France during four years [47]—some of these were described in other countries [69]—and in the strain O12, a clone which caused a different type of non-CF infection in different countries during different years [39]. These observations may indicate that non-CF P. aeruginosa strains could also adapt to human tissues.
Additionally, adaptive evolution of bacteria to a new niche may involve antagonistic pleiotropy [70]; a mutation that is adaptive in the new environment could be detrimental in the original environment [71] and may account for many additional variations found in these strains. In vitro studies indicate that P. aeruginosa derived from CF patients are outcompeted by environmental P. aeruginosa [56, 72], which means that selective pressures during the host colonization make P. aeruginosa less capable of competing in the environment outside the host and that these strains are possibly transmitted from person to person [27].
CONCLUSIONS
The environment outside the host may contain a variety of ecosystems with characteristics that allow the selection of bacterial lineages to colonize human tissue and evade the immune attack. The environment outside the host is also an endless source of opportunistic pathogens, which can infect immunocompromised patients. Controlling and preventing P. aeruginosa infections in hospitals are very complex endeavors that may require an understanding of P. aeruginosa population genetics of the strain causing the outbreak and its relationship with the environment. It is critical to recognize the physiological diversity of the different strains of P. aeruginosa causing infections. Current information suggests that the routes of transmission of the opportunistic pathogen P. aeruginosa are diverse: inanimate objects (such as plumbing and fixtures), human carriers (intestinal or respiratory tract), plants, animals, rivers, etc. It is imperative to use molecular tools to establish whether isolates obtained from different patients or during different timeframes are clonal; genomic information may allow exploring the presence of this pathogen in human carriers, inanimate objects, water sources, etc. Control and preventive measures should be diverse and in agreement with the strain’s origin. Sometimes these measures may require the use of disinfectants or antibiotic treatment of carriers; in other cases, they may require the removal of plumbing or fixtures or other sources of organic matter or water sources.
ACKNOWLEDGMENTS
This research was funded by Universidad San Francisco de Quito USFQ.
AUTHOR CONTRIBUTIONS
Both authors wrote and reviewed the manuscript.
CONFLICT OF INTEREST
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
REFERENCES
[1] Juan, C., Peña, C., & Oliver, A. (2017). Host and Pathogen Biomarkers for Severe Pseudomonas aeruginosa Infections. The Journal of Infectious Diseases, 215(suppl_1), S44-S51. doi: https://doi.org/10.1093/infdis/jiw299
[2] Ding, C., Yang, Z., Wang, J., Liu, X., Cao, Y., Pan, Y., Han, L., & Zhan, S. (2016). Prevalence of Pseudomonas aeruginosa and antimicrobial-resistant Pseudomonas aeruginosa in patients with pneumonia in mainland China: A systematic review and meta-analysis. International Journal of Infectious Diseases, 49, 119-128. doi: https://doi.org/10.1016/j.ijid.2016.06.014
[3] Ribeiro, Á. C. da S., Crozatti, M. T. L., Silva, A. A. da, Macedo, R. S., Machado, A. M. de O., & Silva, A. T. de A. (2020). Pseudomonas aeruginosa in the ICU: Prevalence, resistance profile, and antimicrobial consumption. Revista da Sociedade Brasileira de Medicina Tropical, 53, e20180498. doi: https://doi.org/10.1590/0037-8682-0498-2018
[4] Monsó, E. (2017). Microbiome in chronic obstructive pulmonary disease. Annals of Translational Medicine, 5(12), 251251. doi: https://doi.org/10.21037/atm.2017.04.20
[5] Rodrigo-Troyano, A., & Sibila, O. (2017). The respiratory threat posed by multidrug resistant Gram-negative bacteria: MDR-GNB in respiratory infections. Respirology, 22.7), 1288-1299. doi: https://doi.org/10.1111/resp.13115
[6] Teweldemedhin, M., Gebreyesus, H., Atsbaha, A. H., Asgedom, S. W., & Saravanan, M. (2017). Bacterial profile of ocular infections: A systematic review. BMC Ophthalmology, 17(1). doi: https://doi.org/10.1186/s12886-017-0612-2
[7] Roser, D. J., Van Den Akker, B., Boase, S., Haas, C. N., Ashbolt, N. J., & Rice, S. A. (2014). Pseudomonas aeruginosa dose response and bathing water infection. Epidemiology and Infection, 142(03), 449-462. doi: https://doi.org/10.1017/S0950268813002690
[8] Moremi, N., Claus, H., Vogel, U., & Mshana, S. E. (2017). Surveillance of surgical site infections by Pseudomonas aeruginosa and strain characterization in Tanzanian hospitals does not provide proof for a role of hospital water plumbing systems in transmission. Antimicrobial Resistance & Infection Control, 6(1), 6-56. doi: https://doi.org/10.1186/s13756-017-0216-x
[9] De Soyza, A., & Winstanley, C. (2018). Pseudomonas aeruginosa and Bronchiectasis. In J. Chalmers, E. Polverino, & S. Aliberti (Eds.), Bronchiectasis (pp. 157-180). Springer International Publishing. Cham. doi: https://doi.org/10.1007/978-3-319-61452-6_12
[10] Engelhart, S., Krizek, L., Glasmacher, A., Fischnaller, E., Marklein, G., & Exner, M. (2002). Pseudomonas aeruginosa outbreak in a haematology-oncology unit associated with contaminated surface cleaning equipment. Journal of Hospital Infection, 52(2), 93-98. doi: https://doi.org/10.1053/jhin.2002.1279
[11] English, E. L., Schutz, K. C., Willsey, G. G., & Wargo, M. J. (2018). Transcriptional Responses of Pseudomonas aeruginosa to Potable Water and Freshwater. Applied and Environmental Microbiology, 84(6). doi: https://doi.org/10.1128/AEM.02350-17
[12] Hahn, M. W., & Hofle, M. G. (2001). Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiology Ecology, 35(2), 113-121. doi: https://doi.org/10.1111/j.1574-6941.2001.tb00794.x
[13] Abalos, A., Viñas, M., Sabaté, J., Manresa, M. A., & Solanas, A. M. (2004). Enhanced Biodegradation of Casablanca Crude Oil by a Microbial Consortium in Presence of a Rhamnolipid Produced by Pseudomonas aeruginosa AT10. Biodegradation, 15(4), 249-260. doi: https://doi.org/10.1023/B:BIOD.0000042915.28757.fb
[14] Benie, C. K. D., Dadié, A., Guessennd, N., N’gbesso-Kouadio, N. A., Kouame, N. D., N’golo, D. C., Aka, S., Dako, E., Dje, K. M., & Dosso, M. (2017). Characterization of virulence potential of Pseudomonas aeruginosa isolated from bovine meat, fresh fish, and smoked fish. European Journal of Microbiology and Immunology, 7(1), 55-64. doi: https://doi.org/10.1556/1886.2016.00039
[15] Das, K., & Mukherjee, A. K. (2007). Crude petroleum-oil biodegradation efficiency of Bacillus subtilis and Pseudomonas aeruginosa strains isolated from a petroleum-oil contaminated soil from North-East India. Bioresource Technology, 98(7), 1339-1345. doi: https://doi.org/10.1016/j.biortech.2006.05.032
[16] Gupta, B., Kunal, Rajor, A., & Kaur, J. (2018). Isolation, Characterisation of Novel Pseudomonas and Enterobacter sp. From Contaminated Soil of Chandigarh for Naphthalene Degradation. In S. K. Ghosh (Ed.), Utilization and Management of Bioresources (pp. 175-186). Springer Singapore. doi: https://doi.org/10.1007/978-981-10-5349-8_17
[17] Kotresha, D., & Vidyasagar, G. M. (2008). Isolation and characterisation of phenol-degrading Pseudomonas aeruginosa MTCC 4996. World Journal of Microbiology and Biotechnology, 24(4), 541-547. doi: https://doi.org/10.1007/s11274-007-9508-2
[18] Tortell, P. D., Maldonado, M. T., & Price, N. M. (1996). The role of heterotrophic bacteria in iron-limited ocean ecosystems. Nature, 383(6598), 330-332. doi: https://doi.org/10.1038/383330a0
[19] Wei, Y.-H., Chou, C.-L., & Chang, J.-S. (2005). Rhamnolipid production by indigenous Pseudomonas aeruginosa J4 originating from petrochemical wastewater. Biochemical Engineering Journal, 27(2), 146-154. doi: https://doi.org/10.1016/j.bej.2005.08.028
[20] Bert, F., Maubec, E., Bruneau, B., Berry, P., & Lambert-Zechovsky, N. (1998). Multi-resistant Pseudomonas aeruginosa outbreak associated with contaminated tap water in a neurosurgery intensive care unit. Journal of Hospital Infection, 39(1), 53-62. doi: https://doi.org/10.1016/S0195-6701(98)90243-2
[21] Meyer, B. (2003). Approaches to prevention, removal and killing of biofilms. International Biodeterioration & Biodegradation, 51(4), 249-253. doi: https://doi.org/10.1016/S0964-8305(03)00047-7
[22] Chatterjee, M., Anju, C., Biswas, L., Anil Kumar, V., Gopi Mohan, C., & Biswas, R. (2016). Antibiotic resistance in Pseudomonas aeruginosa and alternative therapeutic options. International Journal of Medical Microbiology: IJMM, 306(1), 48-58. doi: https://doi.org/10.1016/j.ijmm.2015.11.004
[23] Ruiz-Garbajosa, P., & Cantón, R. (2017). Epidemiology of antibiotic resistance in Pseudomonas aeruginosa. Implications for empiric and definitive therapy. Revista Espanola De Quimioterapia, 30 Suppl 1, 8-12. doi: https://doi.org/10.13039/100004325
[24] Subedi, D., Vijay, A. K., & Willcox, M. (2018). Overview of mechanisms of antibiotic resistance in Pseudomonas aeruginosa: An ocular perspective: Mechanism of antimicrobial resistance in P. aeruginosa. Clinical and Experimental Optometry, 101(2), 162-171. doi: https://doi.org/10.1111/cxo.12621
[25] Sokurenko, E. V, Gomulkiewicz, R., & Dykhuizen, D. E. (2006). Source-sink dynamics of virulence evolution. Nature Reviews Microbiology, 4(7), 548-555. doi: https://doi.org/10.1038/nrmicro1446
[26] Dettman, J. R., Rodrigue, N., Aaron, S. D., & Kassen, R. (2013). Evolutionary genomics of epidemic and nonepidemic strains of Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences, 110(52), 21065-21070. doi: https://doi.org/10.1073/pnas.1307862110
[27] Dettman, Jeremy R., Rodrigue, N., & Kassen, R. (2015). Genome-Wide Patterns of Recombination in the Opportunistic Human Pathogen Pseudomonas aeruginosa. Genome Biology and Evolution, 7(1), 18-34. doi: https://doi.org/10.1093/gbe/evu260
[28] Pukatzki, S., Kessin, R. H., & Mekalanos, J. J. (2002). The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proceedings of the National Academy of Sciences of the United States of America, 99(5), 3159-3164. doi: https://doi.org/10.1073/pnas.052704399
[29] Wolfgang, M. C., Kulasekara, B. R., Liang, X., Boyd, D., Wu, K., Yang, Q., Miyada, C. G., & Lory, S. (2003). Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences, 100(14), 8484-8489. doi: https://doi.org/10.1073/pnas.0832438100
[30] Abd, H., Wretlind, B., Saeed, A., Idsund, E., Hultenby, K., & Sandstrá-M, G. (2008). Pseudomonas aeruginosa Utilises Its Type III Secretion System to Kill the Free-Living Amoeba Acanthamoeba castellanii. Journal of Eukaryotic Microbiology, 55(3), 235-243. doi: https://doi.org/10.1111/j.1550-7408.2008.00311.x
[31] Matz, C., Moreno, A. M., Alhede, M., Manefield, M., Hauser, A. R., Givskov, M., & Kjelleberg, S. (2008). Pseudomonas aeruginosa uses type III secretion system to kill biofilm-associated amoebae. The ISME Journal, 2(8), 843-852. doi: https://doi.org/10.1038/ismej.2008.47
[32] Hauser, A. R. (2009). The type III secretion system of Pseudomonas aeruginosa: Infection by injection. Nature Reviews Microbiology, 7(9), 654-665. doi: https://doi.org/10.1038/nrmicro2199
[33] Epelman, S., Stack, D., Bell, C., Wong, E., Neely, G. G., Krutzik, S., Miyake, K., Kubes, P., Zbytnuik, L. D., Ma, L. L., Xie, X., Woods, D. E., & Mody, C. H. (2004). Different Domains of Pseudomonas aeruginosa Exoenzyme S Activate Distinct TLRs. The Journal of Immunology, 173(3), 2031-2040. https://doi.org/10.4049/jimmunol.173.3.2031
[34] Teitzel, G. M., Geddie, A., De Long, S. K., Kirisits, M. J., Whiteley, M., & Parsek, M. R. (2006). Survival and Growth in the Presence of Elevated Copper: Transcriptional Profiling of Copper-Stressed Pseudomonas aeruginosa. Journal of Bacteriology, 188(20), 7242-7256. doi: https://doi.org/10.1128/JB.00837-06
[35] Schalk, I. J., & Guillon, L. (2013). Pyoverdine biosynthesis and secretion in Pseudomonas aeruginosa: Implications for metal homeostasis: Pyoverdine biosynthesis. Environmental Microbiology, 15(6), 1661-1673. doi: https://doi.org/10.1111/1462-2920.12013
[36] Meyer, J. M., Neely, A., Stintzi, A., Georges, C., & Holder, I. A. (1996). Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infection and Immunity, 64(2), 518-523. doi: https://doi.org/10.1128/iai.64.2.518-523.1996
[37] Touati, D. (2000). Iron and Oxidative Stress in Bacteria. Archives of Biochemistry and Biophysics, 373(1), 1-6. doi: https://doi.org/10.1006/abbi.1999.1518
[38] Silby, M. W., Winstanley, C., Godfrey, S. A. C., Levy, S. B., & Jackson, R. W. (2011). Pseudomonas genomes: Diverse and adaptable. FEMS Microbiology Reviews, 35(4), 652-680. doi: https://doi.org/10.1111/j.1574-6976.2011.00269.x
[39] Pirnay, J.-P., Bilocq, F., Pot, B., Cornelis, P., Zizi, M., Van Eldere, J., Deschaght, P., Vaneechoutte, M., Jennes, S., Pitt, T., & De Vos, D. (2009). Pseudomonas aeruginosa Population Structure Revisited. PLoS ONE, 4(11), e7740. doi: https://doi.org/10.1371/journal.pone.0007740
[40] Parcell, B. J., Oravcova, K., Pinheiro, M., Holden, M. T. G., Phillips, G., Turton, J. F., & Gillespie, S. H. (2018). Pseudomonas aeruginosa intensive care unit outbreak: Winnowing of transmissions with molecular and genomic typing. Journal of Hospital Infection, 98(3), 282-288. doi: https://doi.org/10.1016/j.jhin.2017.12.005
[41] Quick, J., Cumley, N., Wearn, C. M., Niebel, M., Constantinidou, C., Thomas, C. M., Pallen, M. J., Moiemen, N. S., Bamford, A., Oppenheim, B., & Loman, N. J. (2014). Seeking the source of Pseudomonas aeruginosa infections in a recently opened hospital: An observational study using whole-genome sequencing. BMJ Open, 4(11), e006278. doi: https://doi.org/10.1136/bmjopen-2014-006278
[42] Sánchez, D., Gomila, M., Bennasar, A., Lalucat, J., & García-Valdés, E. (2014). Genome Analysis of Environmental and Clinical Pseudomonas aeruginosa Isolates from Sequence Type-1146. PLoS ONE, 9(10), e107754. doi: https://doi.org/10.1371/journal.pone.0107754
[43] Lucchetti-Miganeh, C., Redelberger, D., Chambonnier, G., Rechenmann, F., Elsen, S., Bordi, C., Jeannot, K., Attrée, I., Plésiat, P., & de Bentzmann, S. (2014). Pseudomonas aeruginosa Genome Evolution in Patients and under the Hospital Environment. Pathogens, 3(2), 309-340. doi: https://doi.org/10.3390/pathogens3020309
[44] Mahenthiralingam, E., Campbell, M. E., & Speert, D. P. (1994). Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infection and Immunity, 62(2), 596-605.
[45] Diaz Caballero, J., Clark, S. T., Coburn, B., Zhang, Y., Wang, P. W., Donaldson, S. L., Tullis, D. E., Yau, Y. C. W., Waters, V. J., Hwang, D. M., & Guttman, D. S. (2015). Selective Sweeps and Parallel Pathoadaptation Drive Pseudomonas aeruginosa Evolution in the Cystic Fibrosis Lung. MBio, 6(5). e00981-15 doi: https://doi.org/10.1128/mBio.00981-15
[46] Tamburini, F. B., Andermann, T. M., Tkachenko, E., Senchyna, F., Banaei, N., & Bhatt, A. S. (2018). Precision identification of diverse bloodstream pathogens in the gut microbiome. Nature Medicine, 24(12), 1809-1814. doi: https://doi.org/10.1038/s41591-018-0202-8
[47] Cholley, P., Thouverez, M., Hocquet, D., van der Mee-Marquet, N., Talon, D., & Bertrand, X. (2011). Most MultidrugResistant Pseudomonas aeruginosa Isolates from Hospitals in Eastern France Belong to a Few Clonal Types. Journal of Clinical Microbiology, 49(7), 2578-2583. doi: https://doi.org/10.1128/JCM.00102-11
[48] Bhagirath, A. Y., Li, Y., Somayajula, D., Dadashi, M., Badr, S., & Duan, K. (2016). Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulmonary Medicine, 16(1). doi: https://doi.org/10.1186/s12890-016-0339-5
[49] Williams, B. J., Dehnbostel, J., & Blackwell, T. S. (2010). Pseudomonas aeruginosa: Host defense in lung diseases. Respirology, 15(7), 1037-1056. doi: https://doi.org/10.1111/j.1440-1843.2010.01819.x
[50] Perron, G. G., Gonzalez, A., & Buckling, A. (2007). Source-sink dynamics shape the evolution of antibiotic resistance and its pleiotropic fitness cost. Proceedings of the Royal Society B: Biological Sciences, 274(1623), 2351-2356. doi: https://doi.org/10.1098/rspb.2007.0640
[51] Holt, R. D., Barfield, M., & Gomulkiewicz, R. (2004). Temporal Variation Can Facilitate Niche Evolution in Harsh Sink Environments. The American Naturalist, 164(2), 187-200. doi: https://doi.org/10.1086/422343
[52] Kassen, R. (2002). The experimental evolution of specialists, generalists, and the maintenance of diversity: Experimental evolution in variable environments. Journal of Evolutionary Biology, 15(2), 173-190. doi: https://doi.org/10.1046/j.1420-9101.2002.00377.x
[53] Juárez-Vázquez, A. L., Edirisinghe, J. N., Verduzco-Castro, E. A., Michalska, K., Wu, C., Noda-García, L., Babnigg, G., Endres, M., Medina-Ruíz, S., Santoyo-Flores, J., Carrillo-Tripp, M., Ton-That, H., Joachimiak, A., Henry, C. S., & Barona- Gómez, F. (2017). Evolution of substrate specificity in a retained enzyme driven by gene loss. eLife, 6. doi: https://doi.org/10.7554/eLife.22679
[54] Zhang, X., Liu, X., Liang, Y., Guo, X., Xiao, Y., Ma, L., Miao, B., Liu, H., Peng, D., Huang, W., Zhang, Y., & Yin, H. (2017). Adaptive Evolution of Extreme Acidophile Sulfobacillus thermosulfidooxidans Potentially Driven by Horizontal Gene Transfer and Gene Loss. Applied and Environmental Microbiology, 83(7). e03098-16. doi: https://doi.org/10.1128/AEM.03098-16
[55] Moran, N. A. (2002). Microbial minimalism: Genome reduction in bacterial pathogens. Cell, 108(5), 583-586. doi: https://doi.org/10.1016/S0092-8674(02)00665-7
[56] Andersen, S. B., Ghoul, M., Griffin, A. S., Petersen, B., Johansen, H. K., & Molin, S. (2017). Diversity, Prevalence, and Longitudinal Occurrence of Type II Toxin-Antitoxin Systems of Pseudomonas aeruginosa Infecting Cystic Fibrosis Lungs. Frontiers in Microbiology, 8, 1108 doi: https://doi.org/10.3389/fmicb.2017.01180
[57] Barth, A. L., & Pitt, T. L. (1996). The high amino-acid content of sputum from cystic fibrosis patients promotes growth of auxotrophic Pseudomonas aeruginosa. Journal of Medical Microbiology, 45(2), 110-119. doi: https://doi.org/10.1099/00222615-45-2-110
[58] Thomas, S. R. (2000). Increased sputum amino acid concentrations and auxotrophy of Pseudomonas aeruginosa in severe cystic fibrosis lung disease. Thorax, 55(9), 795-797. doi: https://doi.org/10.1136/thorax.55.9.795
[59] Huse, H. K., Kwon, T., Zlosnik, J. E. A., Speert, D. P., Marcotte, E. M., & Whiteley, M. (2010). Parallel Evolution in Pseudomonas aeruginosa over 39,000 Generations In Vivo. MBio, 1(4). doi: https://doi.org/10.1128/mBio.00199-10
[60] Nguyen, A. T., O’Neill, M. J., Watts, A. M., Robson, C. L., Lamont, I. L., Wilks, A., & Oglesby-Sherrouse, A. G. (2014). Adaptation of Iron Homeostasis Pathways by a Pseudomonas aeruginosa Pyoverdine Mutant in the Cystic Fibrosis Lung. Journal of Bacteriology, 196(12), 2265-2276. doi: https://doi.org/10.1128/JB.01491-14
[61] Ghio, A. J., Roggli, V. L., Soukup, J. M., Richards, J. H., Randell, S. H., & Muhlebach, M. S. (2013). Iron accumulates in the lavage and explanted lungs of cystic fibrosis patients. Journal of Cystic Fibrosis, 12(4), 390-398. doi: https://doi.org/10.1016/j.jcf.2012.10.010
[62] Wegiel, B., Nemeth, Z., Correa-Costa, M., Bulmer, A. C., & Otterbein, L. E. (2014). Heme Oxygenase-1: A Metabolic Nike. Antioxidants & Redox Signaling, 20(11), 1709-1722. doi: https://doi.org/10.1089/ars.2013.5667
[63] Huse, H. K., Kwon, T., Zlosnik, J. E. A., Speert, D. P., Marcotte, E. M., & Whiteley, M. (2013). Pseudomonas aeruginosa Enhances Production of a Non-Alginate Exopolysaccharide during Long-Term Colonization of the Cystic Fibrosis Lung. PLoS ONE, 8(12), e82621. doi: https://doi.org/10.1371/journal.pone.0082621
[64] Smith, E. E., Buckley, D. G., Wu, Z., Saenphimmachak, C., Hoffman, L. R., D’Argenio, D. A., Miller, S. I., Ramsey, B. W., Speert, D. P., Moskowitz, S. M., Burns, J. L., Kaul, R., & Olson, M. V. (2006). Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proceedings of the National Academy of Sciences, 103(22), 84878492. doi: https://doi.org/10.1073/pnas.0602138103
[65] Oliver, A., Cantón, R., Campo, P., Baquero, F., & Blázquez, J. (2000). High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science, 288(5469), 1251-1254. doi: https://doi.org/10.1126/science.288.5469.1251
[66] Mena, A., Smith, E. E., Burns, J. L., Speert, D. P., Moskowitz, S. M., Perez, J. L., & Oliver, A. (2008). Genetic Adaptation of Pseudomonas aeruginosa to the Airways of Cystic Fibrosis Patients Is Catalyzed by Hypermutation. Journal of Bacteriology, 190(24), 7910-7917. doi: https://doi.org/10.1128/JB.01147-08
[67] Lam, J., Chan, R., Lam, K., & Costerton, J. W. (1980). Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infection and Immunity, 28(2), 546-556. doi: https://doi.org/10.1128/iai.28.2.546-556.1980
[68] Sriramulu, D. D. (2005). Microcolony formation: A novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. Journal of Medical Microbiology, 54(7), 667-676. doi: https://doi.org/10.1099/jmm.0.45969-0
[69] Treepong, P., Kos, V. N., Guyeux, C., Blanc, D. S., Bertrand, X., Valot, B., & Hocquet, D. (2018). Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clinical Microbiology and Infection, 24(3), 258-266. doi: https://doi.org/10.1016/j.cmi.2017.06.018
[70] Yang, L., Jelsbak, L., Marvig, R. L., Damkiaer, S., Workman, C. T., Rau, M. H., Hansen, S. K., Folkesson, A., Johansen, H. K., Ciofu, O., Hoiby, N., Sommer, M. O. A., & Molin, S. (2011). Evolutionary dynamics of bacteria in a human host environment. Proceedings of the National Academy of Sciences, 108(18), 7481-7486. doi: https://doi.org/10.1073/pnas.1018249108
[71] Kawecki, T. J., & Ebert, D. (2004). Conceptual issues in local adaptation. Ecology Letters, 7(12), 1225-1241. doi: https://doi.org/10.1111/j.1461-0248.2004.00684.x
[72] Chatterjee, P., Davis, E., Yu, F., James, S., Wildschutte, J. H., Wiegmann, D. D., Sherman, D. H., McKay, R. M., LiPuma, J. J., & Wildschutte, H. (2017). Environmental Pseudomonads Inhibit Cystic Fibrosis Patient-Derived Pseudomonas aeruginosa. Applied and Environmental Microbiology, 83(2). doi: https://doi.org/10.1128/AEM.02701-16