Induced mutations in crop plants: mutants of scientific and/or agronomic interest at the Institute of Genetics “Ewald A. Favret”
Mutaciones inducidas en plantas cultivadas: mutantes de interés interés científico y/o agronómico en el Instituto de Genética “Ewald A. Favret”
ACI Avances en Ciencias e Ingenierías
Universidad San Francisco de Quito, Ecuador
Received: 10 August 2020
Accepted: 04 November 2020
Abstract: The use of induced mutations techniques in crop plants at the Institute of Genetics “Ewald A. Favret” (IGEAF) INTA, started in 1949, with the pioneer work of Ewald Favret, who studied the effects of physical and chemical mutagens on barley (Hordeum vulgare) and wheat (Triticum aestivum). IGEAF contributed with several novel results about the effects of important chemical mutagens such as ethyl methane sulfonate (EMS) and sodium azide, and their interactions with X-rays, on barley and wheat. During several decades, a good deal of the research was directed to study the relationship between the different effects of mutagenic treatments on the M. and subsequent generations, and its implications for efficient selection of induced mutants. Many original barley and wheat mutants have been isolated at IGEAF, which early on contributed to elucidate the genetic basis of characters like the hormonal control of growth, the grain protein content and diseases reactions. Besides, several other novel mutants were isolated and characterized including genetically unstable mutants, which are able to originate new heritable variability. One of these mutants is the barley chloroplast mutator (cpm) from which some interesting mutants have been isolated. Moreover, a high throughput strategy for the screening of plastome mutants originated by the cpm was developed (cpTILLING) that allowed the detection of 61 different mutational events, showing the cpm as an extraordinary source of plastome mutants. Furthermore, a mutant allele of the ahas (acetolactate synthase) gene in wheat conferring imidazolinones herbicides tolerance, was isolated. The incorporation of this allele to other genetic backgrounds showed increased levels of tolerance, which in one family were observed in association with increased Fusarium tolerance. In addition to the work done in barley and wheat, interactions with several breeding programs in other crops were carried out. Finally, some commercial achievements of INTA obtained by using induced mutations techniques are briefly described; being the most important the case of the INTA rice (Oryza sativa) breeding program for developing imidazolinones tolerant commercial varieties that in recent years covered 70% of the irrigated rice area in Latin America.
Keywords: Barley mutant, Cotton mutant, Herbicide tolerance, Induced mutations, Mutator genes, Rice mutant, Wheat mutant.
Resumen: El uso de técnicas de mutaciones inducidas en plantas en el Instituto de Genética “Ewald A. Favret” (IGEAF) INTA, se inició en 1949, con el trabajo pionero de Ewald Favret, quien estudió los efectos de mutágenos físicos y químicos en cebada (Hordeum vulgare) y trigo (Triticum aestivum). El IGEAF contribuyó con varios resultados novedosos sobre los efectos de importantes mutágenos químicos como el metanosulfonato de etilo (EMS) y la azida sódica, y sus interacciones con los rayos X, en la cebada y el trigo. Durante varias décadas, gran parte de la investigación se dirigió a estudiar la relación entre los diferentes efectos de los tratamientos mutagénicos en la M. y las generaciones posteriores, y sus implicaciones para la selección eficiente de mutantes inducidos. En el IGEAF se han aislado muchos mutantes originales de cebada y trigo, que desde el principio contribuyeron a interpretar la base genética de caracteres, como el control hormonal del crecimiento, el contenido de proteína del grano y las reacciones a enfermedades. Además, se aislaron y caracterizaron varios otros mutantes nuevos, incluidos mutantes genéticamente inestables, que son capaces de originar una nueva variabilidad heredable. Uno de estos mutantes es el mutador de cloroplastos de la cebada (cpm) del que se han aislado algunos mutantes interesantes. Además, se desarrolló una estrategia de alto rendimiento para la búsqueda de mutantes de cloroplasto originados por cpm (cpTILLING) que permitió la detección de 61 eventos mutacionales diferentes, mostrando a cpm como una fuente extraordinaria de mutantes de cloroplasto. Además, se aisló un alelo mutante del gen ahas (acetolactato sintasa) en trigo que confiere tolerancia a herbicidas imidazolinonas. La incorporación de este alelo a otros acervos genéticos, mostró mayores niveles de tolerancia, que en una familia se observaron en asociación con una mayor tolerancia a Fusarium. Además del trabajo realizado en cebada y trigo, se realizaron interacciones con varios programas de mejoramiento en otros cultivos. Finalmente, se describen brevemente algunos logros comerciales de INTA obtenidos mediante el uso de técnicas de mutaciones inducidas; siendo el caso más importante el del programa de mejoramiento del arroz INTA (Oryza sativa) para el desarrollo de variedades comerciales tolerantes a imidazolinonas, que en los últimos años cubrió el 70% de la superficie arrocera irrigada en América Latina.
Palabras clave: Genes mutadores, Mutante de algodón, Mutante de arroz, Mutante de cebada, Mutaciones inducidas, Mutante de trigo, Tolerancia a herbicidas.
INTRODUCTION
Spontaneous mutations, have facilitated the domestication of several crops, such as, the abolishment of bitterness and toxicity in almonds, and the loss of natural seed dispersal mechanism in peas, barley, wheat, etc. [1]. The concept of mutations in living organisms was established at the beginnings of the last century by Hugo de Vries to designate drastic inheritable changes occasionally occurring in nature. Interestingly, back then de Vries proposed that the rate of spontaneous mutations could be artificially augmented and even be useful in breeding. Indeed, in the 20s, Stadler proved the genetic effects of ionizing radiations on several cultivated plants (barley, maize, wheat and oat) and developed the basis of the experimental mutagenesis in plants. The first commercial success came in 1936 with the release of the first cultivar obtained by using X-rays treatments on tobacco [2, 3, 4].
Since then, these techniques have been used for plant breeding and basic research in many crops either with chemical or physical mutagens. Nowadays, there are more than 3,200 mutant cultivars officially released in more than 220 crops worldwide (https://mvd.iaea.org/). In this review, we summarize the most important achievements regarding induced mutants in different crops obtained within the Institute of Genetics and in collaboration with other breeding programs of INTA.
INDUCED MUTATIONS BACKGROUND AT INSTITUTE OF GENETICS “EWALD A. FAVRET” (IGEAF)
Investigations about induced mutations techniques in crop plants at IGEAF started as early as 1949 by Ewald A. Favret. The institute played a pioneer work in Latin America, contributing with several novel results about the effects of important chemical mutagens such as ethyl methane sulfonate (EMS) and sodium azide and their interactions with X-rays on barley and wheat [5, 6, 7, 8, 9, 10]. During several decades, a good deal of investigations was focused on the study of the relationship between the different effects of mutagenic treatments on the M1 and subsequent generations, and its implications for an efficient selection of induced mutants [3, 4, 11]. From a huge amount of mutagenic treatments many original barley and wheat mutants have been isolated at IGEAF that early on contributed to elucidate the genetic basis of characters like the hormonal control of growth [12], the grain protein content [13], and diseases reactions [6, 14, 15]. In the last decades, several other novel mutants were isolated at IGEAF, which are still under investigation. Some of them are described below.
MUTANTS OF SCIENTIFIC AND OR BREEDING INTEREST
1) DNI (Diverse Number of Internodes) barley mutant
This mutant was isolated after combined treatments of X-rays and sodium azide and it is characterized by showing an extremely high variation in internode numbers among plants and also among different tillers of the same plant, thus it was named DNI. When DNI plants were grown at the field nursery the proportions between normal and multinoded tillers depended on the sowing date. Interestingly, under diverse environmental conditions, the mutant showed several other abnormalities such as, multibranched culms and stolonifer habit, hair like organs emerging from the coleoptile, tillers notably retarding leaf senescence and spikes showing vegetative sectors together with normal ones (see Fig. 1). The gene responsible for DNI phenotype is not known yet; it is hypothesized that it is involved in a very basic mechanism in charge of the establishment and maintenance of meristem identity [16].

FIGURE 1.
DNI barley mutant. A. Plant harvested at the field nursery showing normal tillers carrying mature spikes (yellow arrow) together with multinoded ones (red arrow) carrying vegetative tops with notably retarded leaves senescence. B: DNI (left) and a wild type barley plant (right) grown at greenhouse during winter (short days) and room temperature. DNI mutant shows multibranched culms and stolonifer habit. C. DNI spikes showing vegetative sectors together with normal spikelets, some of them can give normal seeds with germination ability. D. Top with intermediate characteristics between spikelets and leaves. E. Stolonifer habit in a tiller of DNI plant. F. Hair like organ emerging from the coleoptile of a DNI seedling.
2) Barley mutant with roots lacking tropic response to submergence
This mutant was identified when grown in hydroponics [17] by the sandwich method [18]. In those particular conditions, mutant roots did not show the growth pattern with windings and turnings usually observed in wild type barley roots before they submerged (Fig. 2). It was determined that this root behavior is controlled by a semi-dominant nuclear gene [17] and that ethylene produced by mutant roots was significantly lower compared to that of wild type roots when grown in the sandwich method [17, 19]. On the other hand, when grown in humid chambers, where roots grew freely without any physical contact, wild type and mutant roots were morphologically similar and they also had a similar ethylene production [19].
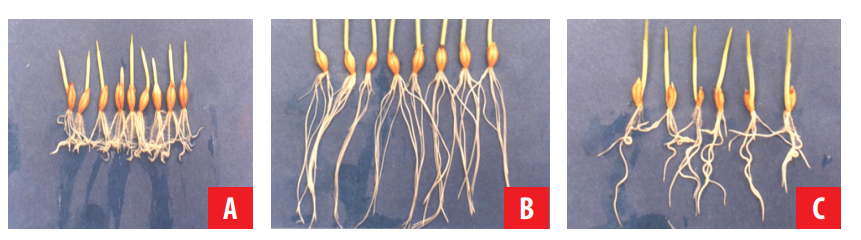
FIGURE 2.
Barley mutant with roots lacking tropic response to submergence. A. Wild type seedlings with roots showing the typical pattern with windings and turnings usually observed when grown in hydroponics by the sandwich method. B. Mutant seedlings lacking tropic response to submergence. C. F1 seedlings showing an intermediate root phenotype.
3) The barley chloroplast mutator mutant (cpm)
The mutator genotype cpm was detected in a family at the M4 generation coming from a combined treatment of X-rays and sodium azide applied on barley seeds. The breeding behavior of the cpm was first described by Prina [20, 21] based on the frequencies of chlorophyll deficiencies, mostly clonally-variegated, observed in seedlings coming from crosses and backcrosses of the cpm with the wild type parental genotype. The cpm originated a wide spectrum of cytoplasmically-inherited chlorophyll deficiencies (Fig. 3). When a cpm plant is crossed as a father with a wild type plant, as plastids are 100% inherited through the ovule in barley, F1 plants are supposed to carry only wild type plastids. As it was expected, F1 plants did not show any chlorophyll deficiencies as evidences of genetic instability. However, chlorophyll deficient narrow streaks appeared in a few seedlings of the F2 generation, and the frequencies of clonally-variegated seedlings increased in the subsequent generations of natural self-pollination [20]. On the other hand, when a plastome mutant is wanted to be genetically stable, the action of the cpm can be stopped using wild type pollen to pollinate the selected plastome mutant and then, in F2 and subsequent generations, selecting plants and families showing solid mutant phenotypes of the selected plastome mutant. The genetically stable plastome mutants achieved in this way were called cytoplasmic lines (CLs), three of which are described below.

FIGURE 3.
The chloroplast mutator mutant (cpm). Some of the chlorophyll deficient phenotypes isolated from cpm plants: A) Virido-xantha seedlings on the left and albino stripped seedlings on the right. B) Viridis seedlings. C) Transversal banded seedlings. D) Albo-viridis seedlings. E) Virido-albino seedlings. F) Highly revertant albino seedlings.
3.1) CL2 mutant
CL2 shows an albo viridis phenotype (Fig. 4), explained as a consequence of a delay in plastid protein synthesis during embryogenesis [22]. The infA gene, which encodes IF1 protein (translation initiation factor 1), was proposed as a candidate gene responsible for CL2 phenotype. When it was sequenced, a missense mutation (T 157 C) was found corresponding to a Ser 52 Pro change in the protein. Other two plastome mutants, showing a similar phenotype to CL2 and independently originated from the original CL2 were isolated (CL2-like 1 and -2). Interestingly, CL2-like 1 also carried a mutation in the infA gene, a T 97 C transition corresponding to a Phe 32 Leu change in the protein [23], while CL2-like 2 presented another different change at the DNA level (A 185 G), corresponding to a change of Asp 61 Gly at IF1 protein. In another experiment, plants of the original genetically stable CL2 mutant were pollinated with cpm pollen, looking for reestablishing the cpm mutation effect on CL2 mutant plastids, which was assumed could be assessed through observing reversions to the wild type color of the CL2 phenotype. One seedling carrying a darker green stripe in the first leaf on a CL2 mutant phenotype background was found and when the infA gene was sequenced, the original CL2 mutation was found plus another missense transition (A 178 G/Met 59 Val) that was interpreted as having a compensation effect on the phenotypic expression of the original CL2 mutant allele [24]. All the above-mentioned results strongly support the relationship between the CL2 phenotype and mutations in the infA gene. It is also worth mentioning that these mutants were the first infA gene mutants described in higher plants.
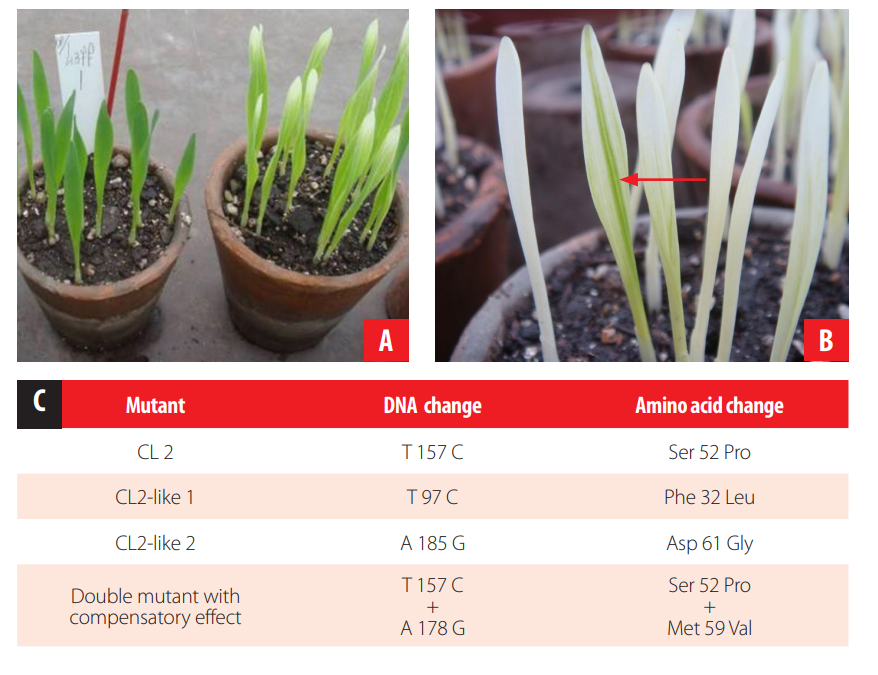
FIGURE 4.
CL2 mutant. A. On the left, wild type barley, on the right the CL2 mutant showing the albo viridis phenotype. B. CL2 seedling showing a phenotypic reversion when carrying the cpm. C. Table showing other infA gene polymorphisms isolated by direct selection of CL2 phenotype reversions independently originated from the CL2 original mutant.
3.2) CL3 mutant
CL3 seedlings were characterized by a homogeneous light green (viridis) phenotype that expressed differentially depending on the temperature (Fig. 5). After several physiological and biochemical analyses, it was concluded that the photosystem I (PSI) was affected but, none of the plastid PSI proteins were altered. However, when ycf3 and ycf4 plastid genes, encoding two chaperones involved in PSI assembly, were sequenced, two mutations were found in intron 1 of ycf3 gene, which affected splicing of this intron at high temperature, and the chaperone encoded by ycf3 was missing. It was concluded that the lack of YCF3 protein during the assembly of PSI at high temperature is the cause of CL3 phenotype [25].
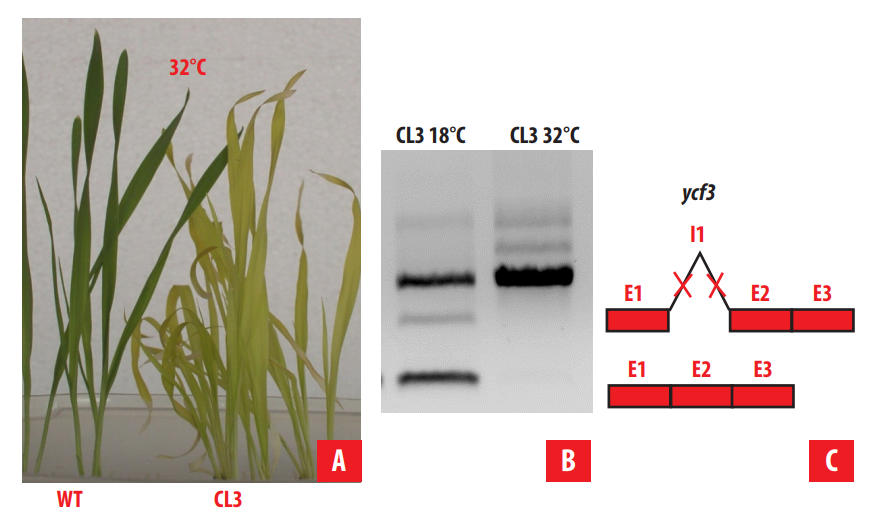
FIGURE 5.
CL3 mutant. A. On the left wild type seedlings, on the right, CL3 seedlings growing at 32°C. B. RT-PCR of ycf3 transcripts at normal and high temperature. C. scheme representing the different transcripts observed in the RT-PCR for CL3 seedlings grown at higher (H) or at low (L) temperatures. The red crosses symbolize the point mutations found in intron 1 (I1) of CL3 that avoid the splicing of I1 under higher temperatures. E: exon, I: intron.
3.3) psbA gene mutant
This mutant was isolated by treating a population carrying the cpm genotype with atrazine, an herbicide toxic for photosystem II (PSII). It was determined that this tolerant mutant carried a mutation in the psbA gene, as it was previously observed to confer atrazine tolerance in several other plant species [26]. The psbA gene encodes D1 protein and atrazine binds to the wild type version of this protein blocking photosynthesis. In field trials this mutant was observed not affected by 8X the recommended dose of atrazine constituting a promising experimental material for developing atrazine tolerant varieties (Fig. 6).

FIGURE 6.
psbA gene mutant. A. Wild type plants without atrazine. B. Wild type plants with 8X atrazine. C. psbA mutant with 8X atrazine.
3.4) cpTILLING (Targeting Induced Local Lesions in Genomes): a high throughput screening strategy for isolation of plastome mutants originated by cpm
The mutants described above were isolated by direct selection based on the phenotype, however, a TILLING strategy was adapted to the chloroplast genome for isolating plastome mutants in cpm populations. TILLING is a reverse genetics high throughput strategy that is very successful for screening nuclear gene mutants of interest in a mutagenized population [27, 28]. For cpTILLING, 31 PCR amplicons were designed comprising 33 genes to analyze 304 cpm seedlings that carried the cpm during different number of generations. At least 61 different mutational events were detected; most of them were transitions and mono nucleotide indels [29]. This strategy also allowed the discovery of a high frequency of seedlings carrying different combinations of five polymorphisms, which already exist in nature, between the rpl23 gene and its pseudogene, which are postulated to originate in an increased rate of illegitimate recombination between these two loci [30]. All the results suggest that the failure of the cpm gene corresponds to a malfunction of a mismatch repair (MMR) gene involved in maintaining the stability of the plastome and that the cpm mutant constitutes an exceptional experimental material to obtain genetic variability in the otherwise highly conserved plastid genome. Cytoplasmic genomes, i.e. those residing in the chloroplast and the mitochondrion, are ruled out by modes of inheritance different than those of the nuclear genome, consequently, the appropriated methodologies for mutation induction and mutants isolation need to be adapted to those circumstances [31].
4) Two cotton (Gossypium hirsutum) mutants of agronomic interest.
In a joint work with the INTA cotton breeding program, located in EEA INTA Sáenz Peña (Chaco Province), a mutant tolerant to imidazolinones was isolated in the M2 generation after a sodium azide mutagenic treatment. Selection was made by applying a solution of imazapir+imazamox (Clearsol Plus BASF). The tolerance was confirmed in subsequent generations indicating it was given by a nuclear gene with semi-dominant expression. The herbicide tolerance was observed to be better expressed after pre-emergency applications, indicating that this mutant would be especially useful in this condition [32]. It was recently determined that the imidazolinone tolerant mutant carries a point mutation in the AHAS gene. Another interesting cotton mutant with pyramidal morphology was isolated after a sodium azide mutagenic treatment. Selection was made on M2 plants grown in glasshouse conditions and later on confirmed in subsequent generations grown at field conditions. The pyramidal morphology is proposed as very useful for narrow furrow harvest (Fig. 7) [33].

FIGURE 7.
Cotton mutant with pyramidal morphology. Cotton plants grown at the greenhouse: A. Wild type. B. Pyramidal mutant showing a much shorter peduncle. C. Schemes of cotton plants after growing in the field nursery: Wild type (left) and pyramidal mutant (right). D. Two mutant plants (left) and a wild type plant (right).
5) A wheat (Triticum aestivum) mutant tolerant to imidazolinones
The search for tolerance to imidazolinone herbicides in wheat was started to cover the need for the development of wheat cultivars tolerant to herbicides with good graminicide characteristics to be used in the South East of Buenos Aires Province. A mutant with tolerance to imidazolinones was isolated from approximately 200,000 M2 seeds of Prointa Elite cv. germinated in a solution of the herbicide. It is well known in bread wheat that carrying ahas mutant alleles in only one of the three genomes, even though it is being used in some commercial cultivars, it gives a somewhat weak level of tolerance. Therefore, most of the imidazolinone tolerant bread wheat commercial varieties carry ahas mutant alleles in two of the three genomes, however, this is associated with a higher fitness cost for the plant. Through cross breeding and backcrosses with several commercial varieties, and selection for increased tolerance to imidazolinones, the tolerance level was increased and moreover, the yield and vigor in three of these families were also increased. Besides, in one of these genetic backgrounds yellow rust and Fusarium tolerance were improved, while in other it was observed a better behavior under high temperature conditions (Fig. 8).
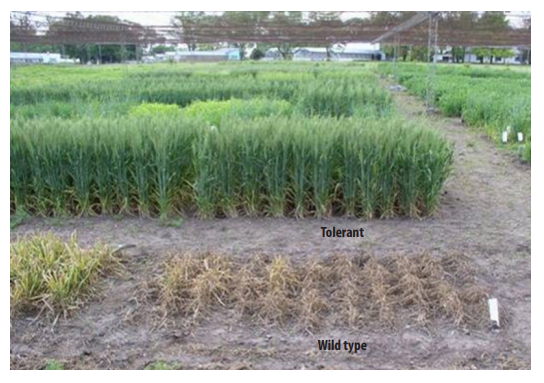
FIGURE 8.
A wheat mutant tolerant to imidazolinones. The mutant and wild type plants were treated with 2X the recommended dose of imazapir solution.
INTA COMMERCIAL ACHIEVEMENTS BY MUTATION BREEDING
The first commercial achievement was obtained in peanut, the mutant variety was named Colorado Irradiado and it came from X-rays irradiated seeds, in a joint work between the IGEAF and EEA INTA Manfredi (Córdoba Province). This mutant was released in 1972 and was characterized for having a high yield and a higher number of fruits. Besides, it was resistant to Cercospora Spp and had a higher oil content. In the 1970s, more than 80% of the peanut cultivated area in Argentina (280,000 ha) was occupied by Colorado Irradiado mutant [34] (https://mvd.iaea.org/#!Variety/144).
Other commercial achievements were the lemon (Citrus limon) cv Eureka 22 INTA, released in 1987, which had a higher fruit set, and the orange (Citrus sinensis) cv Valencia 2 INTA. These mutants were obtained in a joint work between IGEAF and EEA INTA Bella Vista (Corrientes Province).
The biggest commercial success was obtained with the rice mutant Puitá-INTA-CL, which was released in Argentina in 2005. This mutant, which is tolerant to imidazolinones, was obtained in a joint work of the INTA rice breeding program led by Dr. Alberto Livore and located in INTA Concepción, Uruguay (Entre Ríos Province) and IGEAF. Imidazolinones are commonly used herbicides that control a wide spectrum of weeds including the red rice, the most important weed constrain for cultivating rice. After a sodium azide treatment on seeds of three cultivars i.e. El Paso 144, IRGA 417 and Don Juan, and selection in M2 on around six million plants with a mix of imidazolinones, five different alleles of the ahas (acetolactate synthase) gene were isolated and two of them were patented [35, 36]. Three of these mutations were not known in rice. The allele iminta 4 (Ala 122 Thr) was used to obtain the first imidazolinone tolerant variety in Argentina. This generated a commercial agreement with BASF company under the Clear Field technology. Afterwards, three more high yield and quality varieties were released in Argentina, i.e. Guri INTA CL in 2011, Ñu Poty INTA CL in 2013 and Memby Pora INTA CL in 2017. Nowadays, the area sown with imidazolinone tolerant INTA rice varieties in Argentina is around 40% (100 000 ha) and in Brazil, the main rice producer in Latin America, rice varieties carrying the iminta 4 allele, covered more than 800,000 h per year. Varieties carrying iminta 4 allele are also sown in Uruguay, Colombia, Chile, Costa Rica, Panamá, Dominican Republic, Nicaragua, and Honduras. During the 2016/2017 and 2017/2018 campaigns, these varieties covered approximately 70% of the irrigated area in Latin America. In Europe, varieties carrying the iminta 4 allele were released in Italy, Greece, Romania, and Portugal.
ACKNOWLEDGEMENTS
We would like to thank Mr. Abel Mario Moglie and Mr. José Cuello for skillful handling of the plant material. The work of all these years was supported by several Research Contracts and ARCAL projects of the International Atomic Energy Agency; PICTs and PICTOS of the Agencia Nacional de Promoción Científica y Tecnológica, Fondo para la Investigación Científica y Tecnológica/Argentina and research projects of INTA (Instituto Nacional de Tecnología Agropecuaria/Argentina).
AUTHOR’S CONTRIBUTIONS
Alejandra Landau conceived, developed and performed the investigations about cpTILLING, CL2 and CL3 mutant, and sequenced the ahas gene of the cotton mutant; Vanina Brizuela and Daniel Díaz performed the experiments and carried out the greenhouse and field observations about the wheat mutants tolerant to imidazolinones and the atrazine tolerant barley mutant; Franco Lencina performed the investigations about cpTILLING; Alicia Martínez conceived, developed and performed the investigations about the barley mutant with roots lacking tropic response to submergence; Mauricio Tcach and Daniel Díaz conceived, developed and performed the investigations about cotton mutants; María Gabriela Pacheco conceived the investigations about cpTILLING and sequenced the AHAS gene of the wheat mutant; Alberto Prina conceived all the investigations, developed the experimental materials and supervised the greenhouse and field experiments. Alejandra Landau and Alberto Prina wrote the manuscript.
REFERENCES
[1] Mba, C. (2013). Induced mutations unleash the potentials of plant genetic resources for food and agriculture. Agronomy, 3, 200-231. DOI: 10.3390/agronomy3010200
[2] Prakken, R. (1959). Induced Mutation. Euphytica, 8, 270-322. https://link.springer.com/artide/10.1007/BF00039371
[3] Prina, A. R. (1989). Consideraciones sobre la aplicación eficiente de la mutagénesis inducida en fitomejoramiento. Mendeliana, 9, 3-48.
[4] Prina, A. R., Landau, A. M., Pacheco, M. G., & Hopp, H. E. (2010). Mutagénesis, TILLING y EcoTILLING. En G. Levitus, V. Echenique, C. Rubinstein, E. Hopp y L. Mroginski (Eds), Biotecnología y Mejoramiento Vegetal II. Argenbio-INTA. http://exa.unne.edu.ar/biologia/fisiologia.vegetal/BiotecnologiayMejoramientovegetalII.pdf
[5] Favret, E. A. (1960). Somatic mutations of four genes for albinism in barley induced by X-rays and ethyl methanesulphonate. Hereditas, 46 (3-4), 622-634. https://doi.org/10.1111/j.1601-5223.1960.tb03105.x
[6] Favret, E. A. (1960). Spontaneous and induced mutations of barley for the reaction to mildew. Hereditas, 46, 20-28. https://doi.org/10.1111/j.1601-5223.1960.tb03077.x
[7] Favret, E. A. (1964). Genetic effects of single and combined treatment of ionizing radiations and ethyl methane-sulphonate on barley seeds. In: Barley Genetics I. Proceedings of the First International Barley Genetics Symposium,Wageningen. Holland. Proc. Wagenungen PUDOC p. 68-81.
[8] Prina, A. R. & Favret, E. A. (1983). Parabolic effect in sodium azide mutagenesis in barley. Hereditas, 98, 89-94. https://doi.org/10.1111/j.1601-5223.1983.tb00583.x
[9] Prina, A. R. & Favret, E. A. (1983). Influence of potassium cyanide on the azide mutagenesis in barley. Hereditas, 98, 253-258. https://doi.org/10.1111/j.1601-5223.1983.tb00603.x
[10] Prina, A. R., Hagberg, A. & Favret, E. A. (1986). Inheritable sterility induced by X-rays and sodium azide in barley. Genética Agraria, 40, 309-320. https://www.researchgate.net/publication/282255023_Inheritable_sterility_induced_by_X-rays_and_sodium_azide_in_barley
[11] Prina, A. R., Landau, A. M., & Pacheco, M. G. (2012). Chimeras and Mutant Gene Transmisión. In: Q.S. Shu, B. P. Forster & H. Nakagawa (Eds), Plant Mutation Breeding and Biotechnology. FAO/IAEA, International Atomic Energy Agency, Vienna, Austria (ISBN 978-1-78064-085-3) pp 181-189. http://www.fao.org/3/a-i2388e.pdf
[12] Martínez, A. H., & Favret, E. A. (1990). Anthocyanin synthesis and lengthening in the first leaf of barley isogenic lines. Plant Science, 71(1), 35-43. https://doi.org/10.1016/0168-9452(90)90066-W
[13] Favret, E. A., Manghers, L., Solari, R., Avila, A., Monesiglio, J. C. (1970). Gene control of protein production in cereal seeds. Conference paper: Improving plant protein by nuclear techniques. Proceedings of a symposium, Vienna. Jointly organized by the IAEA and FAO. pp.87-97. https://www.cabdirect.org/cabdirect/abstract/19711604439
[14] Favret, E. A. (1965). Induced mutations in breeding for disease resistance. The use of induced mutations in plant breeding. Report of the meeting organised by the FAO/IAEA, 25th may-1st june 1964. Roma. Pergamon. Oxford. GB pp. 521536.
[15] Favret, E. A., Saione, H., Franzone, P. M., Arias, M. C., Solari, R. M. & Lind, V. (1983). Disease reaction mutagenesis in barley and wheat. In: Induced Mutations for Disease resistance in Crop Plants. STI/PUB/633, IAEA, Vienna pp. 53-63.
[16] Prina, A. R. (1995). A developmental barley mutant with temperature conditioned expression and vegetative and sexual reproduction. In: Induced Mutations and Molecular Techniques for Crop Improvement, Proceedings of a symposium, Vienna, 19-23 June 1995, jointly organized by IAEA and FAO, pp 633-634
[17] Martínez, A. E., Franzone, P. M., Aguinaga, A., Polenta, G., Murray, R. & Prina, A. R. (2004). A nuclear gene controlling seminal root growth response to hydroponic cultivation in barley. Environmental and Experimental Botany, 51, 133144. https://doi.org/10.1016/j.envexpbot.2003.09.001
[18] Myhill, R. R. & Konzak, C.F. (1967). A new technique for culturing and measuring barley seedlings. Crop Science, 7, 275-277. https://doi.org/10.2135/cropsci1967.0011183X000700030038x
[19] Martínez, A.E., Landau, A., García, P. T., Polenta, G., Arias, M. C., Murray, R., Pensel, N. & Prina, A. R. (2005). Two mutants affecting adaptative responses to abiotic stresses in barley seedlings. Czech Journal of Genetics and Plant Breeding, 41(1), 1-10. https://www.agriculturejournals.cz/web/cjgpb.htm?type=issue&volume=41&issue=No1
[20] Prina, A. R. (1992). A mutator nuclear gene inducing a wide spectrum of cytoplasmically inherited chlorophyll deficiences in barley. Theoretical and Applied Genetics, 85, 245-251. https://link.springer.com/article/10.1007/BF00222866
[21] Prina, A. R. (1996). Mutator-induced cytoplasmic mutants in barley: genetic evidence of activation of a putative chloroplast transposon. Journal of Heredity, 87, 385-389. https://doi.org/10.1093/oxfordjournals.jhered.a023020
[22] Prina, A. R., Arias, M. C., Lainez, V., Landau, A. & Maldonado, S. (2003). A cytoplasmically inherited mutant controlling early chloroplast development in barley seedlings. Theoretical and Applied Genetics, 107, 1410-1418. https://doi.org/10.1007/s00122-003-1391-0
[23] Landau, A., Díaz Paleo, A., Civitillo, R., Jaureguialzo M., & Prina, A. R. (2007). Two infA gene mutations independently originated from a mutator genotype in barley. Journal of Heredity, 98(3), 272-276. https://doi.org/10.1093/jhered/esm014
[24] Landau, A. M., Pacheco, M. G., & Prina, A. R. (2011). A second infA plastid gene point mutation shows a compensatory effect on the expression of the cytoplasmic line 2 (CL2) syndrome in barley. Journal of Heredity, 102(5), 633-639. https://doi.org/10.1093/jhered/esr061
[25] Landau, A. M., Lokstein, H., Scheller, H. V., Lainez, V., Maldonado, S., & Prina, A.R. (2009). A cytoplasmically inherited barley mutant is defective in photosystem I assembly due to a temperature-sensitive defect in ycf3 splicing. Plant Physiology, 151, 1802-1811. https://doi.org/10.1104/pp.109.147843
[26] Ríos, R.D., Saione, H., Robredo, C., Acevedo, A., Colombo, N., & Prina, A.R. (2003). Isolation and molecular characterization of atrazine tolerant barley mutants. Theoretical and Applied Genetics, 106, 696-702. https://doi.org/10.1007/s00122-002-1119-6
[27] McCallum, C.M., Comai, L., Greene, E.A., & Henikoff, S. (2000). Targeting Induced Local Lesions In Genomes (TILLING) for plant functional genomics. Plant Physiology, 123, 439-442. https://doi.org/10.1104/pp.123.2.439
[28] Colbert, T., Till, B.J., Tompa, R., Reynolds, S., Steine, M.N., Yeung, A.T., McCallum, C.M., Greene, E.A., Comai, L., & Henikoff, S. (2001). High throughtput screening for induced point mutations. Plant Physiology, 126, 480-484. https://doi.org/10.1104/pp.126.2.480.
[29] Landau, A., Lencina, F., Pacheco, M. G., & Prina, A. R. (2016). Plastome mutations and recombination events in barley chloroplast mutator seedlings. Journal of Heredity, 107(3), 266-273. https://doi.org/10.1093/jhered/esw003
[30] Lencina, F., Landau, A. M., Petterson, M. E., Pacheco, M. G., Kobayashi, K., & Prina, A. R. (2019). The rpl23 gene and pseudogene are hotspots of illegitimate recombination in barley chloroplast mutator seedlings. Scientific Reports, 9:9960. https://doi.org/10.1038/s41598-019-46321-6.
[31] Prina, A. R., Pacheco, M. G., & Landau, A. M. (2012) Mutation Induction in Cytoplasmic Genomes. In: Q.S. Shu, B. P. Forster & H. Nakagawa (Eds), Plant Mutation Breeding and Biotechnology. FAO/IAEA, International Atomic Energy Agency, Vienna, Austria (ISBN 978-1-78064-085-3) pp 203-208. http://www.fao.org/3/a-i2388e.pdf
[32] Tcach, M. A., Díaz, D. G., Martínez, A. E., Brizuela, V. E., Acuña, C., & Prina, A. R. (2018). Caracterización de una nueva fuente de tolerancia a imidazolinonas en algodón Gossypium hirsutum obtenida mediante inducción de mutaciones. 1° Congreso Internacional de Algodón, 27/10/17, Presidencia Roque Sáenz Peña, Chaco, Argentina. https://inta.gob.ar/documentos/caracterizacion-de-una-nueva-fuente-de-tolerancia-a-imidazolinonas-en-algodon
[33] Diaz, D. G., Tcach, M., Peterlin, O., Mondino, M., & Prina, A. R. (2011). Selección de líneas de algodón con fructificación agrupada en progenies obtenidas a partir de mutaciones inducidas en Guazuncho 2 INTA. En: Ciencia y Tecnología de los Cultivos Industriales 1 (2) :124-127.
[34] Kharkwal, M. C., & Shu, Q. Y. (2009). The Role of Induced Mutations in World Food Security. In: Q.Y. Shu (Ed), Induced Plant Mutations in the Genomics Era. FAO/IAEA, Rome, Italy. (ISBN 978-92-5-106324-8) pp 33-38. https://mvd.iaea.org/PDF/IPM200901.pdf
[35] Livore, A. B., Prina, A. R., Singh, B. K., Ascenzi, R., & Whitt, S. R. (2010). Herbicide-resistant rice plants, polynucleotides encoding herbicide-resistant acetohydroxyacid synthase large subunit proteins, and methods of use. United States Patent application publication. https://patents.justia.com/inventor/alberto-livore
[36] Livore, A. B., Prina, A. R., Birk, I., & Singh, B. (2011). Rice plants having increased tolerance to imidazolinone herbicides. European patent application. https://patents.justia.com/inventor/alberto-livore